Note
Go to the end to download the full example code.
Interpolating Datasets#
Interpolate slices from standard PSI data files.
- This example demonstrates how to use
Read in the metadata for a 3D data file (the radial magnetic field data).
br_data_filepath = data.get_3d_data()
metadata = read_hdf_meta(br_data_filepath)
Datasets for file: br.h5
+-----------------+-----------------+-----------------+
| Name | Shape | Type |
+-----------------+-----------------+-----------------+
| Data | (181, 100, 151) | float32 |
+-----------------+-----------------+-----------------+
Scales for file: br.h5
+------------+------------+------------+------------+------------+------------+------------+
| Dataset | Dim | Name | Shape | Type | Min | Max |
+------------+------------+------------+------------+------------+------------+------------+
| Data | 0 | dim1 | (151,) | float32 | 0.99956024 | 30.511646 |
| Data | 1 | dim2 | (100,) | float32 | 0.0 | 3.1415927 |
| Data | 2 | dim3 | (181,) | float32 | 0.0 | 6.2831855 |
+------------+------------+------------+------------+------------+------------+------------+
Using this metadata, we can see that the dataset is a standard PSI 3D data file with scales:
radius (1st dimension), in units of solar radii (R☉),
co-latitude (2nd dimension), in radians,
longitude (3rd dimension), in radians.
Suppose we wish to interpolate the given dataset at a specific radial value,
say at 30 R☉, across all latitudes and longitudes. The most efficient approach is
through the function np_interpolate_slice_from_hdf(), which uses
vectorized NumPy computation for interpolation, while also leveraging
read_hdf_by_value() to only read in the minimum amount of data
to perform these computations.
data_at_30 = np_interpolate_slice_from_hdf(br_data_filepath, 30, None, None)
for r in data_at_30:
print(r.shape)
(181, 100)
(100,)
(181,)
Note
np_interpolate_slice_from_hdf() results in a dimensional
reduction of the dataset. In this case, the original 3D dataset is reduced to a 2D
slice at the specified radial value.
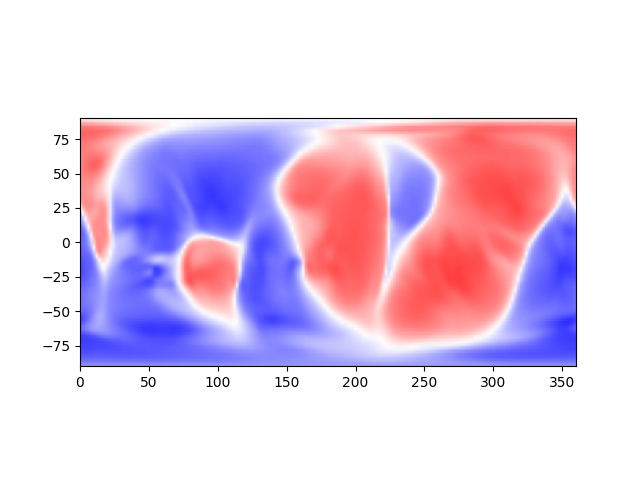
Given the metadata fetched above, we can visualize this slice as a long-lat map:
data, scale_t, scale_p = data_at_30
ax = plt.figure().add_subplot()
ax.pcolormesh(np.rad2deg(scale_p),
90-np.rad2deg(scale_t),
data.T,
cmap='bwr',
shading='gouraud',
clim=(-5e-4,5e-4))
ax.set_aspect("equal", adjustable="box")
plt.show()
Alternatively, if we wish to interpolate the dataset at arbitrary
(radius, co-latitude, longitude) positions, we can use
interpolate_positions_from_hdf(). For example, to interpolate
at some (contrived) trajectory:
r_positions = np.linspace(1.0, 30.0, 10) # from 1 R☉ to 30 R☉
theta_positions = np.linspace(0.0, np.pi, 10) # from
phi_positions = np.linspace(0.0, 2*np.pi, 10) # from 0 to 2π
interpolated_values = interpolate_positions_from_hdf(
br_data_filepath,
r_positions,
theta_positions,
phi_positions
)
for value, position in zip(interpolated_values, zip(r_positions, theta_positions, phi_positions)):
print(f"(radius={position[0]:.2f} R☉, "
f"latitude={90-np.rad2deg(position[1]):.2f}°, "
f"longitude={np.rad2deg(position[2]):.2f}°) ->\n"
f" B_r = {value:.4e} MAS UNITS")
(radius=1.00 R☉, latitude=90.00°, longitude=0.00°) ->
B_r = 3.3264e-01 MAS UNITS
(radius=4.22 R☉, latitude=70.00°, longitude=40.00°) ->
B_r = -2.4614e-03 MAS UNITS
(radius=7.44 R☉, latitude=50.00°, longitude=80.00°) ->
B_r = -4.8336e-03 MAS UNITS
(radius=10.67 R☉, latitude=30.00°, longitude=120.00°) ->
B_r = -2.4804e-03 MAS UNITS
(radius=13.89 R☉, latitude=10.00°, longitude=160.00°) ->
B_r = 7.3004e-04 MAS UNITS
(radius=17.11 R☉, latitude=-10.00°, longitude=200.00°) ->
B_r = 1.0084e-03 MAS UNITS
(radius=20.33 R☉, latitude=-30.00°, longitude=240.00°) ->
B_r = 6.2016e-04 MAS UNITS
(radius=23.56 R☉, latitude=-50.00°, longitude=280.00°) ->
B_r = 3.9668e-04 MAS UNITS
(radius=26.78 R☉, latitude=-70.00°, longitude=320.00°) ->
B_r = -2.3550e-04 MAS UNITS
(radius=30.00 R☉, latitude=-90.00°, longitude=360.00°) ->
B_r = -1.9969e-04 MAS UNITS
Total running time of the script: (0 minutes 0.156 seconds)
