Getting started¶
psipy is a package for loading and visualising the output of PSI’s MAS model runs. This page provides some narrative documentation that should get you up and running with obtaining, loading, and visualising some model results.
Installing¶
Currently the only way to install psipy is from source. Change to the directory you want to download the source too, and run:
git clone https://github.com/predsci/PsiPy
cd PsiPy
pip install .
Getting data¶
The PSI MHDWeb pages give access to MAS model runs. The runs are indexed by Carrington rotation, and for each Carrington rotation there are generally a number of different runs, varying in the type model run and/or the boundary conditions.
To load data with psipy you need to manually download the files you are interested in to a directory on your computer.
Loading data¶
psipy stores the output variables from a single MAS run in the
MASOutput object. To create one of these, specify the directory
which has all of the outputs .hdf files you want to load:
from psipy.model import MASOutput
directory = '/path/to/files'
mas_output = MASOutput(directory)
To see which variables have been loaded, we can look at the .variables
attribute:
print(mas_output.variables)
# TODO: PUT OUTPUT HERE
This will print a list of the variables that have been loaded. Each individual variable can then be accessed with square brackets, for example to get the radial magnetic field component:
br = mas_output['br']
print(br)
# TODO: PUT OUTPUT HERE
This will return a Variable object, which stores the underlying data as a
xarray.DataArray under Variable.data.
Data coordinates¶
The data stored in Variable.data contains values of the data as a normal
array, but in addition also stores the coordinates of each data point.
MAS model outputs are defined on a 3D grid of points on a spherical grid. The
coordinate names are 'r', 'theta', 'phi'.
TODO: explain how to get coordinates
Plotting data¶
A MASOutput instance can be indexed to get individual Variable data, e.g.:
br = mas_output['br']
These Variable objects have methods to plot 2D slices of the data. These
methods are:
A typical use looks like this:
ax = plt.subplot(1, 1, 1, projection='polar')
model['rho'].plot_phi_cut(index, ax=ax, ...)
and produces an output like this:
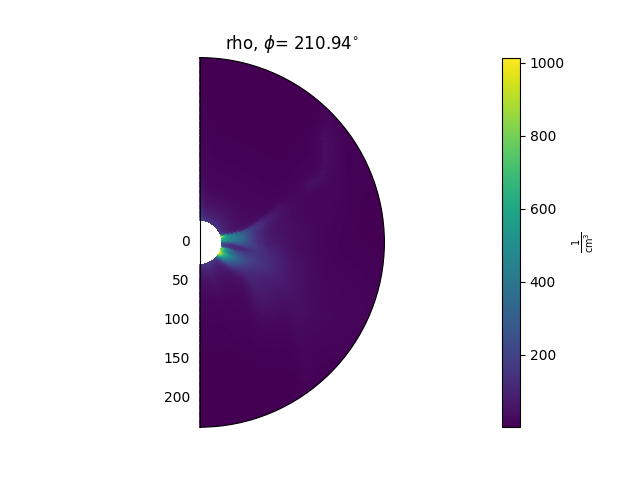
For more examples of how to use these methods, see the Examples gallery.
There are also methods that can be used to plot contours of the data on top of these 2D slices. As an example, this can be helpful for plotting the heliospheric current sheet, by contouring \(B_{r} = 0\). These methods are
A typical use looks like this:
ax = plt.subplot(1, 1, 1, projection='polar')
model['rho'].plot_phi_cut(index, ax=ax, ...)
model['br'].contour_phi_cut(index, levels=[0], ax=ax, ...)
and produces outputs like this:
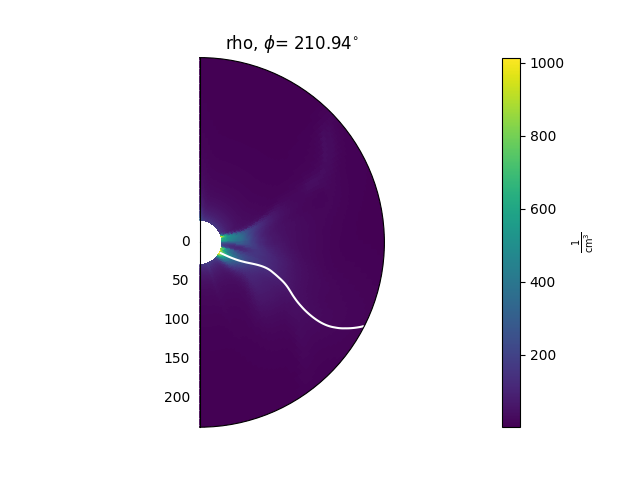
For more examples of how to use these methods, see the Examples gallery.
Normalising data before plotting¶
Sometimes it is helpful to multiply data by an expected radial falloff, e.g.
multiplying the density by \(r^{2}\). This can be done using the
Variable.radial_normalized method, e.g.:
rho = mas_output['rho']
rho_r_squared = rho.radial_normalized(-2)
rho_r_squared.plot_phi_cut(...)